T he Glass laboratory investigates transcriptional mechanisms that regulate the development and function of the macrophage, a cell that plays key roles in immunity and inflammatory diseases. Current efforts are to determine the biochemical and biological roles of sequence-specific transcription factors and their associated co-regulators at gene-specific and genome-wide scales. A combination of biochemical, cellular and genetic model systems are used, incorporating macrophage-specific knockouts, microarray technologies, massively parallel sequencing and bioinformatics approaches, to unravel the contributions of specific factors to the development of specialized macrophage functions in immunity and the pathogenesis of inflammatory diseases.
Research Interests
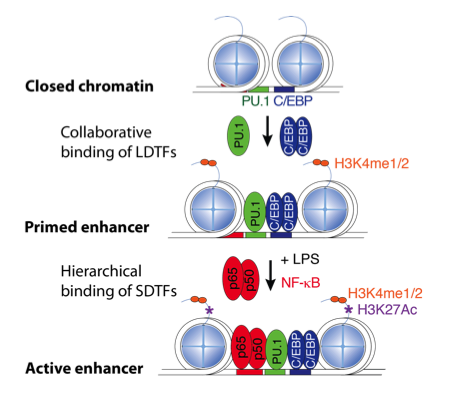
Enhancer Selection
and Functions
We are investigating mechanisms by which macrophage lineage determining factors, such as PU.1, prime enhancers for subsequent actions of signal dependent transcription factors, such as NFkB and members of the nuclear receptor superfamily.
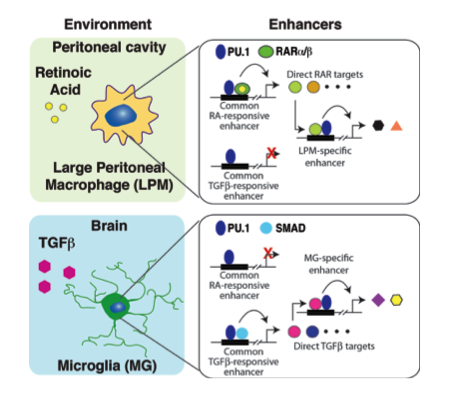
Macrophage
subtypes
We are using genome-wide approaches to define the mechanisms by which developmental origin and tissue environment determine mouse and human macrophage functions in health and disease.
Genetic
variation
We are exploiting natural genetic variation provided by different inbred strains of mice as a ‘mutagenesis strategy’ to define transcription factor networks in macrophages. These approaches are being extended to understand the effects of natural genetic variation on disease phenotypes in humans.
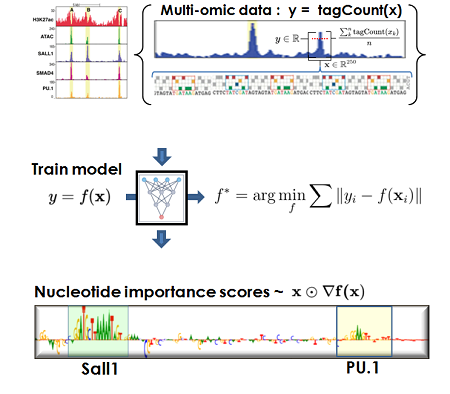
Machine Learning
guided discovery
We are developing novel machine learning (ML) formulations for capturing models from multi-omic data corresponding to the immune system cells, macrophages. We use ML models for structure discovery of transcription factor (TF) compositions and networks, in silico mutagenesis, cell fate and signal driven enhancer selection and function.


